Identification of an aphid resistance gene in Arabidopsis
Karen Kloth used EntoLab in her research on the genetic basis of plant resistance to aphids. Genome-wide association mapping of aphid behavior on 350 natural Arabidopsis accessions led to the discovery of a new gene.
Posted by
Published on
Sun 20 Sep. 2020
Topics
| Behavioral Research | Insects | EntoLab |

Aphids are a huge problem in farming systems, as they transfer plant viruses and drain precious nutrients and sugars from crops. Plants have a plethora of natural defense mechanisms against these herbivorous insects, but crops are often devoid of them and extra vulnerable as they are grown in monocultures. Therefore, insecticides are applied on a large scale. Now the use of many insecticidal compounds is becoming restricted due to their detrimental impact on the environment, there is an increasing demand to breed crops that have natural defense mechanisms against aphids. But for this, the genetic basis of plant resistance to aphids needs to be known. In a quest to identify plant resistance genes, we were faced with the vast task of screening hundreds of Arabidopsis plant lines for resistance to aphids.
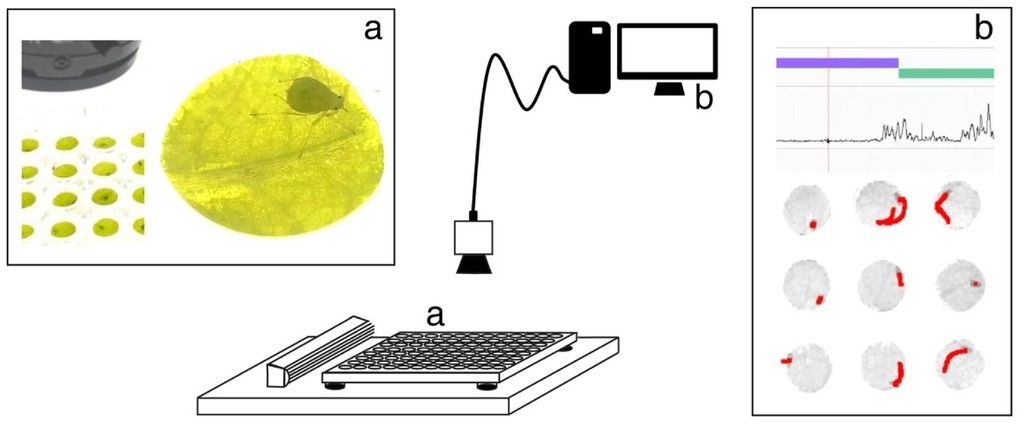
Initial setup for video tracking used by Kloth et al. (2015). In the current EntoLab assay plates, larger leaf segments are used to maintain better phloem turgor, and up to 300 instead of 20 arenas can be imaged. Water condensation issues have been solved allowing prolonged measurements and the detection of additional (induced) phenotypes.
With conventional techniques this would require a big greenhouse space and months of manually counting aphid larvae and adults that usually hide in the tiniest spaces underneath the leaves. Instead, we used automated video tracking as a high-throughput phenotyping method to quantify aphid feeding behavior. In collaboration with Noldus Information Technology, we developed a video-tracking platform with a top-view camera above a well plate with aphids on leaf discs. With EthoVision software, aphid body movements were automatically tracked. With this, we could calculate the number of ‘probes’ the aphids performed, i.e. the punctures aphids make in the leaf with their piercing-sucking mouthparts. From the duration of each probe we could estimate which probes were unsuccessful (short, superficial penetrations) and which probes had a high likelihood of involving successful feeding events (long probes) and determine the level of resistance of each plant. Eventually, we were able to screen 6 biological replicates of each of the 350 plant lines in a couple of weeks with this prototype platform (which later became the EntoLab system). Not only did it save us a lot of time and manual labor, we also obtained relevant details about different aspects of plant resistance, quantified in more than 20 behavioral variables. With genome-wide association mapping, one of these variables revealed a significant association with mutations in a gene on chromosome 3 of Arabidopsis.
Movie of automated video tracking of 20 aphids simultaneously. Myzus persicae aphids were tracked on Arabidopsis leaf discs using EthoVision XT video tracking software. The aphid’s body center was automatically detected and tracks of the previous 30 seconds are visualized in red. Arenas are numbered from 1 (top left) to 20 (bottom right). The movie is displayed at 16× speed.
Follow-up experiments on this gene - with a thus far unknown function - showed that it encodes a small heat shock-like protein that localizes in the phloem and reduces phloem feeding and Myzus persicae aphid reproduction with around 50% in a time frame of two weeks. We named this gene SIEVE ELEMENT-LINING CHAPERONE1 (SLI1). In recent work, we found that SLI1 is also involved in quantitative plant resistance to the cabbage aphid, Brevicoryne brassicae, and the cabbage whitefly, Aleyrodes proletella.
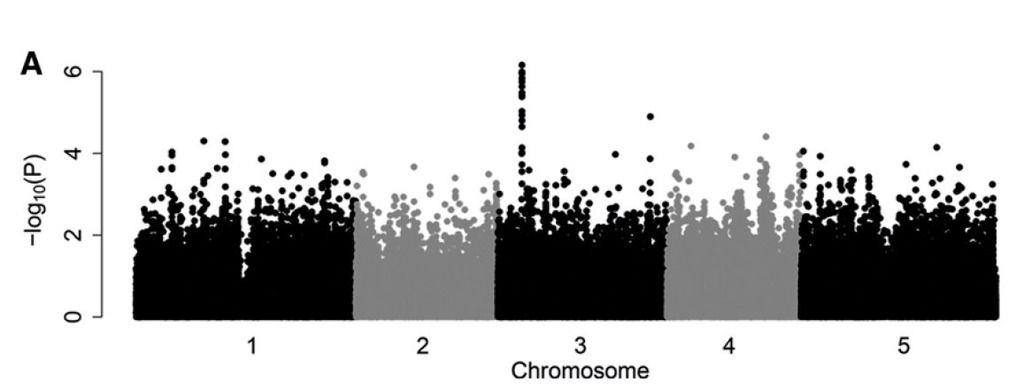
GWA mapping of Myzus persicae aphid feeding behavior. Genome-wide associations of the total time aphids spent on short probes (<3 min, n = 3–6, 350 accessions, 214 k SNPs). A conventional screening for population growth did not yield any association.
References
- Farquharson, K. (2017). A phloem protein contributes to aphid resistance and heat stress tolerance. Blog, The Plant Cell: In Brief, 6 Oct 2017. https://plantae.org/a-phloem-protein-contributes-to-aphid-resistance-and-heat-stress-tolerance/
- Kloth, K.J. (2016). Mapping moves on Arabidopsis: From natural variation to single genes affecting aphid behaviour. PhD thesis, Wageningen University, 272 pp. https://edepot.wur.nl/370445
- Kloth, K.J.; ten Broeke, C.J.M.; Thoen, M.P.M.; Den Brink, M.H.V.; Wiegers, G.L.; Krips, O.E.; Noldus, L.P.J.J.; Dicke, M.; Jongsma, M.A. (2015). High-throughput phenotyping of plant resistance to aphids by automated video tracking. Plant Methods, 11:4. http://dx.doi.org/10.1186/s13007-015-0044-z
- Kloth, K.J.; Busscher-Lange, J.; Wiegers, G.L.; Kruijer, W.; Buijs, G.; Meyer, R.C.; Bouwmeester, H.J.; Dicke, M.; Jongsma, M.A. (2017). Sieve element-Lining Chaperone 1 restricts aphid feeding on Arabidopsis during heat stress. The Plant Cell, 29:2450-2464. http://dx.doi.org/10.1105/tpc.16.00424
- Kloth, K.J.; Wiegers, G.L.; Busscher-Lange, J.; van Haarst, J.C.; Kruijer, W.; Bouwmeester, H.J.; Dicke, M.; Jongsma, M.A. (2016). WRKY22 promotes susceptibility to aphids and modulates salicylic acid and jasmonic acid signalling. Journal of Experimental Botany, 67, 2283-3396. http://dx.doi.org/10.1093/jxb/erw159
- Kloth, K.J.; Shah, P.; Broekgaarden, C.; Ström, C.; Albrectsen, B.R.; Dicke, M. (2021). SLI1 confers broad-spectrum resistance to phloem-feeding insects. Plant, Cell & Environment. https://doi.org/10.1111/pce.14064
Related Posts
The value of tracking software: using EthoVision XT to monitor broiler activity

What can you expect from Noldus at SfN 2023?
